2A.ml - Statistiques descriptives avec scikit-learn - correction#
Links: notebook, html, python, slides, GitHub
ACP, CAH, régression linéaire, correction.
%matplotlib inline
import matplotlib.pyplot as plt
from jyquickhelper import add_notebook_menu
add_notebook_menu()
# Répare une incompatibilité entre scipy 1.0 et statsmodels 0.8.
from pymyinstall.fix import fix_scipy10_for_statsmodels08
fix_scipy10_for_statsmodels08()
Prérequis de l’énoncé#
import pyensae.datasource
f = pyensae.datasource.download_data("dads2011_gf_salaries11_dbase.zip",
website="https://www.insee.fr/fr/statistiques/fichier/2011542/")
import pandas
try:
from dbfread import DBF
use_dbfread = True
except ImportError as e :
use_dbfread = False
if use_dbfread:
import os
from pyensae.sql.database_exception import ExceptionSQL
from pyensae.datasource import dBase2sqllite
print("convert dbase into sqllite")
try:
dBase2sqllite("salaries2011.db3", "varlist_salaries11.dbf", overwrite_table="varlist")
dBase2sqllite("salaries2011.db3", "varmod_salaries11.dbf", overwrite_table="varmod")
dBase2sqllite("salaries2011.db3", 'salaries11.dbf', overwrite_table="salaries", fLOG = print)
except ExceptionSQL:
print("La base de données est déjà renseignée.")
else :
print("use of zipped version")
import pyensae.datasource
db3 = pyensae.datasource.download_data("salaries2011.zip")
# pour aller plus vite, données à télécharger au
# http://www.xavierdupre.fr/enseignement/complements/salaries2011.zip
convert dbase into sqllite
unable to execute a SQL request (1)(file salaries2011.db3)
create table varlist (VARIABLE TEXT, LIBELLE TEXT, TYPE TEXT, LONGUEUR REAL)
La base de données est déjà renseignée.
Exercice 1 : CAH (classification ascendante hiérarchique)#
Le point commun de ces méthodes est qu’elles ne sont pas supervisées. L’objectif est de réduire la complexité des données. Réduire le nombre de dimensions pour l’ACP ou segmenter les observations pour les k-means et la CAH.
import pandas, numpy, matplotlib.pyplot as plt
from sklearn.decomposition import PCA
import pyensae.datasource
pyensae.datasource.download_data("eleve_region.txt")
df = pandas.read_csv("eleve_region.txt", sep="\t", encoding="utf8", index_col=0)
print(df.shape)
df.head(n=5)
(27, 21)
| 1993 | 1994 | 1995 | 1996 | 1997 | 1998 | 1999 | 2000 | 2001 | 2002 | ... | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| académie | |||||||||||||||||||||
| Aix-Marseille | 241357 | 242298 | 242096 | 242295 | 243660 | 244608 | 245536 | 247288 | 249331 | 250871 | ... | 250622 | 248208 | 245755 | 243832 | 242309 | 240664 | 240432 | 241336 | 239051 | 240115 |
| Amiens | 198281 | 196871 | 195709 | 194055 | 192893 | 191862 | 189636 | 185977 | 183357 | 180973 | ... | 175610 | 172110 | 168718 | 165295 | 163116 | 162548 | 163270 | 164422 | 165275 | 166345 |
| Besançon | 116373 | 115600 | 114282 | 113312 | 112076 | 110261 | 108106 | 105463 | 103336 | 102264 | ... | 100117 | 98611 | 97038 | 95779 | 95074 | 94501 | 94599 | 94745 | 94351 | 94613 |
| Bordeaux | 253551 | 252644 | 249658 | 247708 | 247499 | 245757 | 244992 | 243047 | 243592 | 245198 | ... | 244805 | 244343 | 242602 | 242933 | 243146 | 244336 | 246806 | 250626 | 252085 | 255761 |
| Caen | 145435 | 144369 | 141883 | 140658 | 139585 | 137704 | 135613 | 133255 | 131206 | 129271 | ... | 125552 | 123889 | 122550 | 121002 | 119857 | 119426 | 119184 | 119764 | 119010 | 119238 |
5 rows × 21 columns
for c in df.columns:
if c != "1993":
df[c] /= df ["1993"]
df["1993"] /= df["1993"]
df.head()
| 1993 | 1994 | 1995 | 1996 | 1997 | 1998 | 1999 | 2000 | 2001 | 2002 | ... | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| académie | |||||||||||||||||||||
| Aix-Marseille | 1.0 | 1.003899 | 1.003062 | 1.003886 | 1.009542 | 1.013470 | 1.017315 | 1.024574 | 1.033038 | 1.039419 | ... | 1.038387 | 1.028385 | 1.018222 | 1.010255 | 1.003944 | 0.997129 | 0.996168 | 0.999913 | 0.990446 | 0.994854 |
| Amiens | 1.0 | 0.992889 | 0.987029 | 0.978687 | 0.972826 | 0.967627 | 0.956400 | 0.937947 | 0.924733 | 0.912710 | ... | 0.885662 | 0.868011 | 0.850904 | 0.833640 | 0.822651 | 0.819786 | 0.823427 | 0.829237 | 0.833539 | 0.838936 |
| Besançon | 1.0 | 0.993358 | 0.982032 | 0.973697 | 0.963076 | 0.947479 | 0.928961 | 0.906250 | 0.887972 | 0.878761 | ... | 0.860311 | 0.847370 | 0.833853 | 0.823035 | 0.816976 | 0.812053 | 0.812895 | 0.814149 | 0.810764 | 0.813015 |
| Bordeaux | 1.0 | 0.996423 | 0.984646 | 0.976955 | 0.976131 | 0.969261 | 0.966243 | 0.958572 | 0.960722 | 0.967056 | ... | 0.965506 | 0.963684 | 0.956817 | 0.958123 | 0.958963 | 0.963656 | 0.973398 | 0.988464 | 0.994218 | 1.008716 |
| Caen | 1.0 | 0.992670 | 0.975577 | 0.967154 | 0.959776 | 0.946842 | 0.932465 | 0.916251 | 0.902162 | 0.888858 | ... | 0.863286 | 0.851851 | 0.842644 | 0.832001 | 0.824128 | 0.821164 | 0.819500 | 0.823488 | 0.818304 | 0.819871 |
5 rows × 21 columns
from sklearn.cluster import AgglomerativeClustering
ward = AgglomerativeClustering(linkage='ward', compute_full_tree=True).fit(df)
ward
AgglomerativeClustering(compute_full_tree=True)
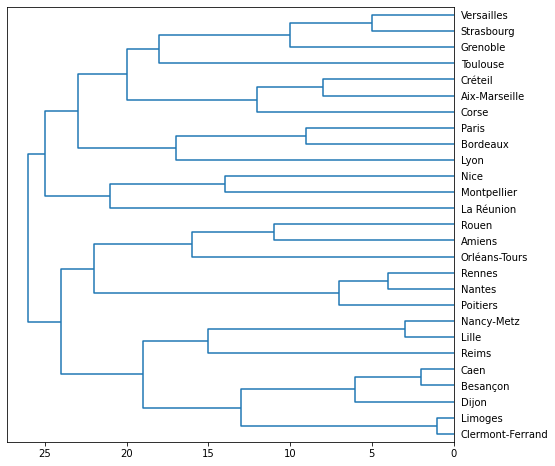
from scipy.cluster.hierarchy import dendrogram
import matplotlib.pyplot as plt
dendro = [ ]
for a,b in ward.children_:
dendro.append([a, b, float(len(dendro)+1), len(dendro)+1])
# le dernier coefficient devrait contenir le nombre de feuilles dépendant de ce noeud
# et non le dernier indice
# de même, le niveau (3ème colonne) ne devrait pas être le nombre de noeud
# mais la distance de Ward
fig = plt.figure( figsize=(8,8) )
ax = fig.add_subplot(1,1,1)
r = dendrogram(dendro, color_threshold=1, labels=list(df.index),
show_leaf_counts=True, ax=ax, orientation="left")
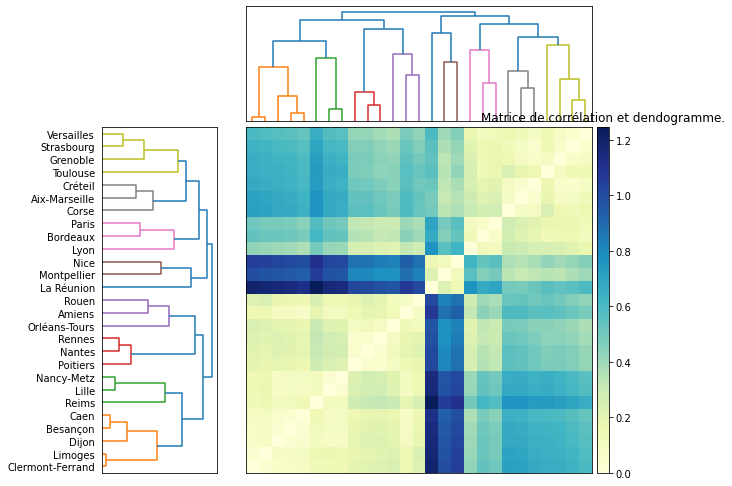
Je reprends également le graphique montrant la matrice de corrélations qu’on peut également obtenir avec seaborn : clustermap.
from scipy.spatial.distance import pdist, squareform
data_dist = pdist(df)
fig = plt.figure(figsize=(8,8))
# x ywidth height
ax1 = fig.add_axes([0.05,0.1,0.2,0.6])
Z1 = dendrogram(dendro, orientation='right',labels=list(df.index))
ax1.set_xticks([])
# Compute and plot second dendrogram.
ax2 = fig.add_axes([0.3,0.71,0.6,0.2])
Z2 = dendrogram(dendro)
ax2.set_xticks([])
ax2.set_yticks([])
# Compute and plot the heatmap
axmatrix = fig.add_axes([0.3,0.1,0.6,0.6])
idx1 = Z1['leaves']
idx2 = Z2['leaves']
D = squareform(data_dist)
D = D[idx1,:]
D = D[:,idx2]
im = axmatrix.matshow(D, aspect='auto', origin='lower', cmap=plt.cm.YlGnBu)
axmatrix.set_xticks([])
axmatrix.set_yticks([])
# Plot colorbar.
axcolor = fig.add_axes([0.91,0.1,0.02,0.6])
plt.colorbar(im, cax=axcolor)
plt.title("Matrice de corrélation et dendogramme.");
Exercice 2 : régression linéaire#
Ce sont trois méthodes supervisées : on s’en sert pour expliquer prédire
le lien entre deux variables et
(ou ensemble de
variables) ou prédire
en fonction de
. On suppose que
les données ont déjà été téléchargées (voir l’énoncé de cet exercice).
On continue avec l’extraction de l’âge, du sexe et du salaire comme
indiqué dans l’énoncé.
import sqlite3, pandas
con = sqlite3.connect("salaries2011.db3")
df = pandas.io.sql.read_sql("select * from varmod", con)
con.close()
values = df[ df.VARIABLE == "TRNNETO"].copy()
def process_intervalle(s):
# [14 000 ; 16 000[ euros
acc = "0123456789;+"
s0 = "".join(c for c in s if c in acc)
spl = s0.split(';')
if len(spl) != 2:
raise ValueError("Unable to process '{0}'".format(s0))
try:
a = float(spl[0])
except Exception as e:
raise ValueError("Cannot interpret '{0}' - {1}".format(s, spl))
b = float(spl[1]) if "+" not in spl[1] else None
if b is None:
return a
return (a+b) / 2.0
values["montant"] = values.apply(lambda r : process_intervalle(r["MODLIBELLE"]), axis=1)
values.head()
| VARIABLE | MODALITE | MODLIBELLE | montant | |
|---|---|---|---|---|
| 8957 | TRNNETO | 00 | [0 ; 200[ euros | 100.0 |
| 8958 | TRNNETO | 01 | [200 ; 500[ euros | 350.0 |
| 8959 | TRNNETO | 02 | [500 ; 1 000[ euros | 750.0 |
| 8960 | TRNNETO | 03 | [1 000 ; 1 500[ euros | 1250.0 |
| 8961 | TRNNETO | 04 | [1 500 ; 2 000[ euros | 1750.0 |
import sqlite3, pandas
con = sqlite3.connect("salaries2011.db3")
data = pandas.io.sql.read_sql("select TRNNETO,AGE,SEXE from salaries", con)
con.close()
salaires = data.merge (values, left_on="TRNNETO", right_on="MODALITE" )
salaires["M"] = salaires.apply(lambda r: 1 if r["SEXE"] == "1" else 0, axis=1)
salaires["F"] = salaires.apply(lambda r: 1 if r["SEXE"] == "2" else 0, axis=1)
data = salaires[["AGE","M","F","montant"]]
data = data [data.M + data.F > 0]
data.head()
| AGE | M | F | montant | |
|---|---|---|---|---|
| 0 | 49.0 | 1 | 0 | 750.0 |
| 1 | 27.0 | 1 | 0 | 750.0 |
| 2 | 22.0 | 1 | 0 | 750.0 |
| 3 | 26.0 | 1 | 0 | 750.0 |
| 4 | 29.0 | 0 | 1 | 750.0 |
On supprime les valeurs manquantes :
nonull = data.dropna().copy()
nonull.shape
(2240691, 4)
version scikit-learn#
La régression linéraire suit :
nonull[["AGE","M"]].dropna().shape
(2240691, 2)
from sklearn import linear_model
clf = linear_model.LinearRegression()
clf.fit (nonull[["AGE","M"]].values, nonull.montant.values)
clf.coef_, clf.intercept_, "R^2=", clf.score(
nonull[["AGE","M"]], nonull.montant)
C:Python395_x64libsite-packagessklearnbase.py:443: UserWarning: X has feature names, but LinearRegression was fitted without feature names warnings.warn(
(array([ 310.98873096, 4710.02901965]),
4927.219006697269,
'R^2=',
0.13957345814666222)
On prend un échantillon aléatoire :
import random
val = nonull.copy()
val["rnd"] = val.apply(lambda r: random.randint(0, 1000), axis=1)
ech = val[val["rnd"] == 1]
ech.shape
(2278, 5)
On sépare homme et femmes :
homme = ech[ech.M == 1]
femme = ech[ech.M == 0]
predh = clf.predict(homme[["AGE","M"]])
predf = clf.predict(femme[["AGE","M"]])
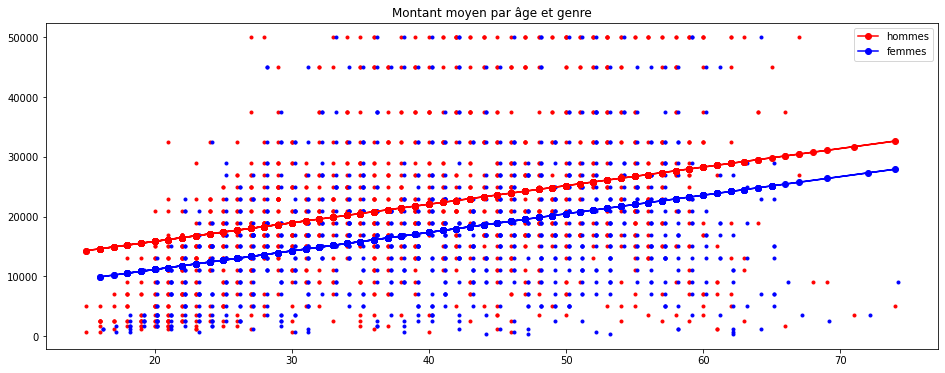
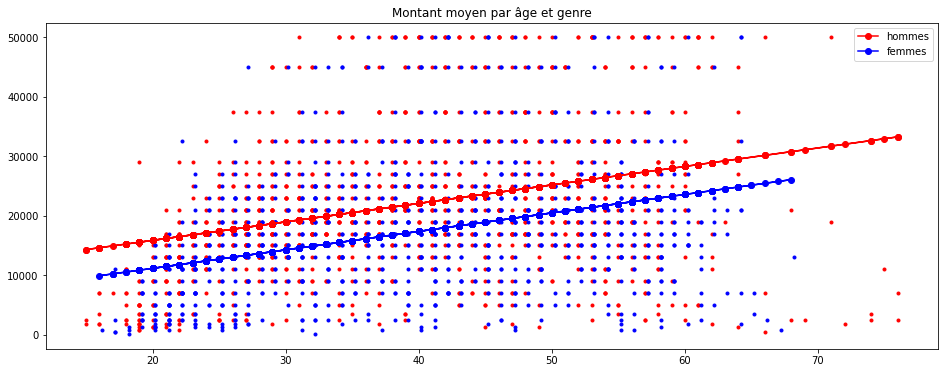
import matplotlib.pyplot as plt
plt.figure(figsize=(16,6))
plt.plot(homme.AGE, homme.montant, "r.")
plt.plot(femme.AGE + 0.2, femme.montant, "b.")
plt.plot(homme.AGE, predh, "ro-", label="hommes")
plt.plot(femme.AGE, predf, "bo-", label="femmes")
plt.legend()
plt.title("Montant moyen par âge et genre");
C:Python395_x64libsite-packagessklearnbase.py:443: UserWarning: X has feature names, but LinearRegression was fitted without feature names warnings.warn( C:Python395_x64libsite-packagessklearnbase.py:443: UserWarning: X has feature names, but LinearRegression was fitted without feature names warnings.warn(
version statsmodels#
import statsmodels.api as sm
nonull["one"] = 1.0 # on ajoute la constante
model = sm.OLS(nonull.montant, nonull [["AGE","M", "one"]])
results = model.fit()
print("coefficients",results.params)
results.summary()
C:Python395_x64libsite-packagesstatsmodelstsabasetsa_model.py:7: FutureWarning: pandas.Int64Index is deprecated and will be removed from pandas in a future version. Use pandas.Index with the appropriate dtype instead. from pandas import (to_datetime, Int64Index, DatetimeIndex, Period, C:Python395_x64libsite-packagesstatsmodelstsabasetsa_model.py:7: FutureWarning: pandas.Float64Index is deprecated and will be removed from pandas in a future version. Use pandas.Index with the appropriate dtype instead. from pandas import (to_datetime, Int64Index, DatetimeIndex, Period,
coefficients AGE 310.988731
M 4710.029020
one 4927.219007
dtype: float64
| Dep. Variable: | montant | R-squared: | 0.140 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.140 |
| Method: | Least Squares | F-statistic: | 1.817e+05 |
| Date: | Sat, 12 Feb 2022 | Prob (F-statistic): | 0.00 |
| Time: | 18:57:08 | Log-Likelihood: | -2.4023e+07 |
| No. Observations: | 2240691 | AIC: | 4.805e+07 |
| Df Residuals: | 2240688 | BIC: | 4.805e+07 |
| Df Model: | 2 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| AGE | 310.9887 | 0.600 | 518.682 | 0.000 | 309.814 | 312.164 |
| M | 4710.0290 | 14.658 | 321.323 | 0.000 | 4681.299 | 4738.759 |
| one | 4927.2190 | 26.032 | 189.272 | 0.000 | 4876.196 | 4978.242 |
| Omnibus: | 132298.944 | Durbin-Watson: | 0.183 |
|---|---|---|---|
| Prob(Omnibus): | 0.000 | Jarque-Bera (JB): | 158649.997 |
| Skew: | 0.614 | Prob(JB): | 0.00 |
| Kurtosis: | 3.440 | Cond. No. | 150. |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
On reproduit le même dessin :
import random
val = nonull.copy()
val["rnd"] = val.apply(lambda r: random.randint(0,1000), axis=1)
ech = val[val["rnd"] == 1]
homme = ech [ ech.M == 1]
femme = ech [ ech.M == 0]
predh = results.predict(homme[["AGE","M","one"]])
predf = results.predict(femme[["AGE","M","one"]])
import matplotlib.pyplot as plt
def graph(homme, femme, predh, predf):
fig, ax = plt.subplots(1, 1, figsize=(16, 6))
ax.plot(homme.AGE, homme.montant, "r.")
ax.plot(femme.AGE + 0.2, femme.montant, "b.")
ax.plot(homme.AGE, predh, "ro-", label="hommes")
ax.plot(femme.AGE, predf, "bo-", label="femmes")
ax.legend()
ax.set_title("Montant moyen par âge et genre");
return ax
graph(homme, femme, predh, predf);
On ajoute l’intervalle de confiance sur un échantillon :
from statsmodels.sandbox.regression.predstd import wls_prediction_std
prstd, iv_l, iv_u = wls_prediction_std(results)
val = nonull.copy()
val["rnd"] = val.apply(lambda r: random.randint(0, 1000), axis=1)
val["pred"] = prstd
val["up"] = iv_u
val["down"] = iv_l
ech = val[val["rnd"] == 1]
ech.head()
| AGE | M | F | montant | one | rnd | pred | up | down | |
|---|---|---|---|---|---|---|---|---|---|
| 86 | 34.0 | 0 | 1 | 750.0 | 1.0 | 1 | 10964.546920 | 36990.964539 | -5989.292820 |
| 355 | 48.0 | 0 | 1 | 750.0 | 1.0 | 1 | 10964.547492 | 41344.807893 | -1635.451707 |
| 1046 | 68.0 | 0 | 1 | 750.0 | 1.0 | 1 | 10964.559457 | 47564.605962 | 4584.299462 |
| 2008 | 21.0 | 0 | 1 | 750.0 | 1.0 | 1 | 10964.552144 | 32948.121273 | -10032.156559 |
| 3761 | 18.0 | 1 | 0 | 750.0 | 1.0 | 1 | 10964.553468 | 36725.186695 | -6255.096328 |
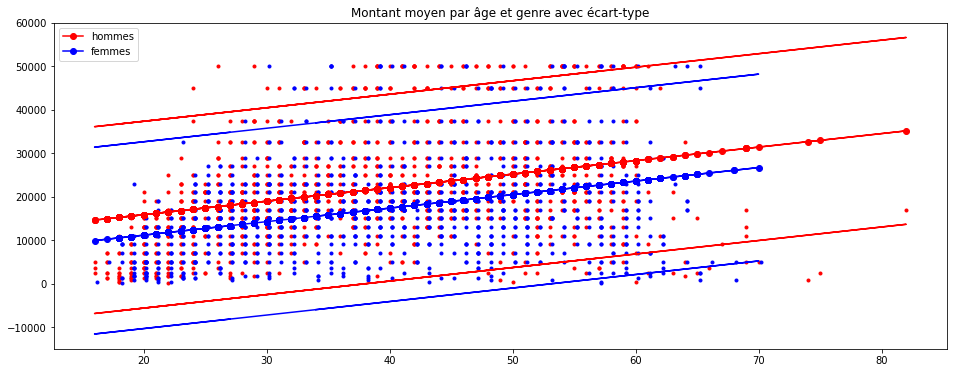
Puis on l’ajoute au graphe précédent :
homme = ech[ech.M == 1]
femme = ech[ech.M == 0]
predh = results.predict(homme[["AGE","M","one"]])
predf = results.predict(femme[["AGE","M","one"]])
ax = graph(homme, femme, predh, predf)
ax.plot(homme.AGE, homme.up, 'r-')
ax.plot(homme.AGE, homme.down, 'r-')
ax.plot(femme.AGE, femme.up, 'b-')
ax.plot(femme.AGE, femme.down, 'b-')
ax.set_title("Montant moyen par âge et genre avec écart-type");