2018-10-09 Ensemble, Gradient, Boosting…#
Links: notebook, html, python, slides, GitHub
Le noteboook explore quelques particularités des algorithmes d’apprentissage pour expliquer certains résultats numériques. L’algoithme AdaBoost surpondère les exemples sur lequel un modèle fait des erreurs.
from jyquickhelper import add_notebook_menu
add_notebook_menu()
%matplotlib inline
Skewed split train test#
Lorsqu’une classe est sous représentée, il est difficile de prédire les résultats d’un modèle de machine learning.
import numpy, numpy.random
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.neural_network import MLPClassifier
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier
from sklearn.metrics import confusion_matrix
N = 1000
res = []
for n in [1, 2, 5, 10, 20, 50, 80, 90, 100, 110]:
print("n=", n)
for k in range(10):
X = numpy.zeros((N, 2))
X[:, 0] = numpy.random.randint(0, 2, (N,))
X[:, 1] = numpy.random.randint(0, n+1, (N,))
Y = X[:, 0] + X[:, 1] + numpy.random.normal(size=(N,)) / 2
Y[Y < 1.5] = 0
Y[Y >= 1.5] = 1
X_train, X_test, y_train, y_test = train_test_split(X, Y)
stat = dict(N=N, n=n, ratio_train=y_train.sum()/y_train.shape[0],
k=k, ratio_test=y_test.sum()/y_test.shape[0])
for model in [LogisticRegression(solver="liblinear"),
MLPClassifier(max_iter=500),
RandomForestClassifier(n_estimators=10),
AdaBoostClassifier(DecisionTreeClassifier(), n_estimators=10)]:
obs = stat.copy()
obs["model"] = model.__class__.__name__
if obs["model"] == "AdaBoostClassifier":
obs["model"] = "AdaB-" + model.base_estimator.__class__.__name__
try:
model.fit(X_train, y_train)
except ValueError as e:
obs["erreur"] = str(e)
res.append(obs)
continue
sc = model.score(X_test, y_test)
obs["accuracy"] = sc
conf = confusion_matrix(y_test, model.predict(X_test))
try:
obs["Error-0|1"] = conf[0, 1] / conf[0, :].sum()
obs["Error-1|0"] = conf[1, 0] / conf[1, :].sum()
except Exception:
pass
res.append(obs)
n= 1
n= 2
n= 5
n= 10
n= 20
n= 50
n= 80
n= 90
n= 100
n= 110
from pandas import DataFrame
df = DataFrame(res)
df = df.sort_values(['n', 'model', 'model', "k"]).reset_index(drop=True)
df["diff_ratio"] = (df["ratio_test"] - df["ratio_train"]).abs()
df.head(n=5)
| N | n | ratio_train | k | ratio_test | model | accuracy | Error-0|1 | Error-1|0 | diff_ratio | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1000 | 1 | 0.273333 | 0 | 0.300 | AdaB-DecisionTreeClassifier | 0.860 | 0.062857 | 0.320000 | 0.026667 |
| 1 | 1000 | 1 | 0.274667 | 1 | 0.328 | AdaB-DecisionTreeClassifier | 0.916 | 0.029762 | 0.195122 | 0.053333 |
| 2 | 1000 | 1 | 0.304000 | 2 | 0.284 | AdaB-DecisionTreeClassifier | 0.860 | 0.072626 | 0.309859 | 0.020000 |
| 3 | 1000 | 1 | 0.285333 | 3 | 0.268 | AdaB-DecisionTreeClassifier | 0.896 | 0.027322 | 0.313433 | 0.017333 |
| 4 | 1000 | 1 | 0.297333 | 4 | 0.256 | AdaB-DecisionTreeClassifier | 0.888 | 0.053763 | 0.281250 | 0.041333 |
df.tail(n=5)
| N | n | ratio_train | k | ratio_test | model | accuracy | Error-0|1 | Error-1|0 | diff_ratio | |
|---|---|---|---|---|---|---|---|---|---|---|
| 395 | 1000 | 110 | 0.982667 | 5 | 0.996 | RandomForestClassifier | 0.996 | 0.0 | 0.004016 | 0.013333 |
| 396 | 1000 | 110 | 0.990667 | 6 | 0.980 | RandomForestClassifier | 0.996 | 0.2 | 0.000000 | 0.010667 |
| 397 | 1000 | 110 | 0.985333 | 7 | 0.988 | RandomForestClassifier | 1.000 | 0.0 | 0.000000 | 0.002667 |
| 398 | 1000 | 110 | 0.985333 | 8 | 0.992 | RandomForestClassifier | 1.000 | 0.0 | 0.000000 | 0.006667 |
| 399 | 1000 | 110 | 0.985333 | 9 | 0.992 | RandomForestClassifier | 0.996 | 0.5 | 0.000000 | 0.006667 |
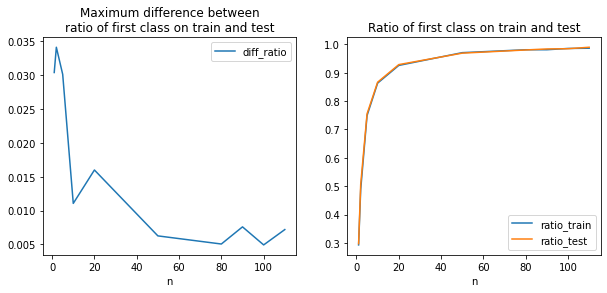
La répartition train/test est loin d’être statisfaisante lorsqu’il existe une classe sous représentée.
df[df.n==100][["n", "ratio_test", "ratio_train"]].head(n=10)
| n | ratio_test | ratio_train | |
|---|---|---|---|
| 320 | 100 | 0.980 | 0.992000 |
| 321 | 100 | 0.984 | 0.980000 |
| 322 | 100 | 0.988 | 0.984000 |
| 323 | 100 | 0.988 | 0.986667 |
| 324 | 100 | 0.976 | 0.986667 |
| 325 | 100 | 0.984 | 0.985333 |
| 326 | 100 | 0.984 | 0.981333 |
| 327 | 100 | 0.988 | 0.982667 |
| 328 | 100 | 0.984 | 0.989333 |
| 329 | 100 | 0.992 | 0.989333 |
#df.to_excel("data.xlsx")
columns = ["n", "N", "model"]
agg = df.groupby(columns, as_index=False).mean().sort_values(["n", "model"]).reset_index(drop=True)
agg.tail()
| n | N | model | ratio_train | k | ratio_test | accuracy | Error-0|1 | Error-1|0 | diff_ratio | |
|---|---|---|---|---|---|---|---|---|---|---|
| 35 | 100 | 1000 | RandomForestClassifier | 0.985733 | 4.5 | 0.9848 | 0.9956 | 0.185000 | 0.001216 | 0.004933 |
| 36 | 110 | 1000 | AdaB-DecisionTreeClassifier | 0.986533 | 4.5 | 0.9900 | 0.9972 | 0.130000 | 0.000810 | 0.007200 |
| 37 | 110 | 1000 | LogisticRegression | 0.986533 | 4.5 | 0.9900 | 0.9960 | 0.346667 | 0.000402 | 0.007200 |
| 38 | 110 | 1000 | MLPClassifier | 0.986533 | 4.5 | 0.9900 | 0.9956 | 0.346667 | 0.000810 | 0.007200 |
| 39 | 110 | 1000 | RandomForestClassifier | 0.986533 | 4.5 | 0.9900 | 0.9980 | 0.090000 | 0.000810 | 0.007200 |
import matplotlib.pyplot as plt
fig, ax = plt.subplots(1, 2, figsize=(10,4))
agg.plot(x="n", y="diff_ratio", ax=ax[0])
agg.plot(x="n", y="ratio_train", ax=ax[1])
agg.plot(x="n", y="ratio_test", ax=ax[1])
ax[0].set_title("Maximum difference between\nratio of first class on train and test")
ax[1].set_title("Ratio of first class on train and test")
ax[0].legend();
Une astuce pour éviter les doublons avant d’effecturer un pivot.
agg2 = agg.copy()
agg2["ratio_test2"] = agg2["ratio_test"] + agg2["n"] / 100000
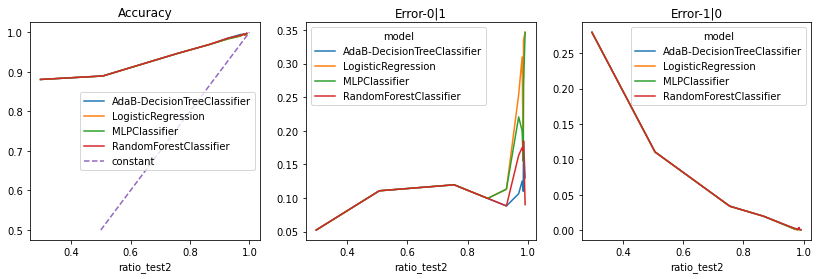
import matplotlib.pyplot as plt
fig, ax = plt.subplots(1, 3, figsize=(14,4))
agg2.pivot("ratio_test2", "model", "accuracy").plot(ax=ax[0])
agg2.pivot("ratio_test2", "model", "Error-0|1").plot(ax=ax[1])
agg2.pivot("ratio_test2", "model", "Error-1|0").plot(ax=ax[2])
ax[0].plot([0.5, 1.0], [0.5, 1.0], '--', label="constant")
ax[0].set_title("Accuracy")
ax[1].set_title("Error-0|1")
ax[2].set_title("Error-1|0")
ax[0].legend();
agg2.pivot("ratio_test2", "model", "Error-0|1")
| model | AdaB-DecisionTreeClassifier | LogisticRegression | MLPClassifier | RandomForestClassifier |
|---|---|---|---|---|
| ratio_test2 | ||||
| 0.29721 | 0.052249 | 0.052249 | 0.052249 | 0.052249 |
| 0.50682 | 0.110686 | 0.110686 | 0.110686 | 0.110686 |
| 0.75525 | 0.119578 | 0.119578 | 0.119578 | 0.119578 |
| 0.86690 | 0.099333 | 0.099333 | 0.099333 | 0.099333 |
| 0.92900 | 0.088095 | 0.113095 | 0.113095 | 0.088095 |
| 0.96970 | 0.106349 | 0.253968 | 0.220635 | 0.163492 |
| 0.98120 | 0.125000 | 0.310000 | 0.200000 | 0.175000 |
| 0.98490 | 0.110000 | 0.155000 | 0.155000 | 0.170000 |
| 0.98580 | 0.185000 | 0.335000 | 0.268333 | 0.185000 |
| 0.99110 | 0.130000 | 0.346667 | 0.346667 | 0.090000 |
Le modèle AdaBoost construit 10 arbres tout comme la forêt aléatoire à ceci près que le poids associé à chacun des arbres des différents et non uniforme.
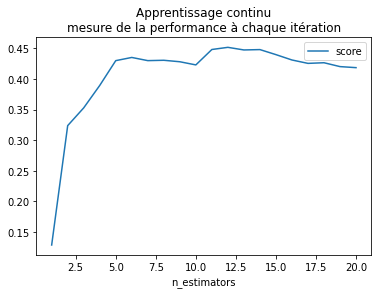
Apprentissage continu#
Apprendre une forêt aléatoire, puis ajouter un arbre, encore un tout en gardant le résultat des apprentissages précédents.
from sklearn.datasets import load_diabetes
data = load_diabetes()
X, y = data.data, data.target
X_train, X_test, y_train, y_test = train_test_split(X, y)
from sklearn.ensemble import RandomForestRegressor
model = None
res = []
for i in range(0, 20):
if model is None:
model = RandomForestRegressor(n_estimators=1, warm_start=True)
else:
model.set_params(**dict(n_estimators=model.n_estimators+1))
model.fit(X_train, y_train)
score = model.score(X_test, y_test)
res.append(dict(n_estimators=model.n_estimators, score=score))
df = DataFrame(res)
df.head()
| n_estimators | score | |
|---|---|---|
| 0 | 1 | 0.128906 |
| 1 | 2 | 0.323854 |
| 2 | 3 | 0.352876 |
| 3 | 4 | 0.389476 |
| 4 | 5 | 0.429992 |
ax = df.plot(x="n_estimators", y="score")
ax.set_title("Apprentissage continu\nmesure de la performance à chaque itération");