ONNX side by side#
Links: notebook, html, PDF, python, slides, GitHub
The notebook compares two runtimes for the same ONNX and looks into differences at each step of the graph.
from jyquickhelper import add_notebook_menu
add_notebook_menu()
%load_ext mlprodict
The mlprodict extension is already loaded. To reload it, use:
%reload_ext mlprodict
%matplotlib inline
The ONNX model#
We convert kernel function used in GaussianProcessRegressor. First some values to use for testing.
import numpy
import pandas
from io import StringIO
Xtest = pandas.read_csv(StringIO("""
1.000000000000000000e+02,1.061277971307766705e+02,1.472195004809226493e+00,2.307125069497626552e-02,4.539948095743629591e-02,2.855191098141335870e-01
1.000000000000000000e+02,9.417031896832908444e+01,1.249743892709246573e+00,2.370416174339620707e-02,2.613847280316268853e-02,5.097165413593484073e-01
1.000000000000000000e+02,9.305231488674536422e+01,1.795726729335217264e+00,2.473274733802270642e-02,1.349765645107412620e-02,9.410288840541443378e-02
1.000000000000000000e+02,7.411264142156210255e+01,1.747723020195752319e+00,1.559695663417645997e-02,4.230394035515055301e-02,2.225492746314280956e-01
1.000000000000000000e+02,9.326006195761877393e+01,1.738860294343326229e+00,2.280160135767652502e-02,4.883335335161764074e-02,2.806808409247734115e-01
1.000000000000000000e+02,8.341529291866362428e+01,5.119682123742423929e-01,2.488795768635816003e-02,4.887573336092913834e-02,1.673462179673477768e-01
1.000000000000000000e+02,1.182436477919874562e+02,1.733516391831658954e+00,1.533520930349476820e-02,3.131213519485807895e-02,1.955345358785769427e-01
1.000000000000000000e+02,1.228982583299257101e+02,1.115599996405831629e+00,1.929354155079938959e-02,3.056996308544096715e-03,1.197052763998271013e-01
1.000000000000000000e+02,1.160303269386108838e+02,1.018627021014927303e+00,2.248784981616459844e-02,2.688111547114307651e-02,3.326105131778724355e-01
1.000000000000000000e+02,1.163414374640396005e+02,6.644299545804077667e-01,1.508088417713602906e-02,4.451836657613789106e-02,3.245643044204808425e-01
""".strip("\n\r ")), header=None).values
Then the kernel.
from sklearn.gaussian_process.kernels import RBF, ConstantKernel as CK, Sum
ker = Sum(
CK(0.1, (1e-3, 1e3)) * RBF(length_scale=10,
length_scale_bounds=(1e-3, 1e3)),
CK(0.1, (1e-3, 1e3)) * RBF(length_scale=1,
length_scale_bounds=(1e-3, 1e3))
)
ker
0.316**2 * RBF(length_scale=10) + 0.316**2 * RBF(length_scale=1)
ker(Xtest)
array([[2.00000000e-01, 4.88993040e-02, 4.25048140e-02, 5.94472678e-04,
4.36813578e-02, 7.54738292e-03, 4.79816083e-02, 2.44870899e-02,
6.11804858e-02, 5.91636643e-02],
[4.88993040e-02, 2.00000000e-01, 1.41439850e-01, 1.33559792e-02,
1.56539930e-01, 5.58967934e-02, 5.50622994e-03, 1.61259456e-03,
9.16550083e-03, 8.54623880e-03],
[4.25048140e-02, 1.41439850e-01, 2.00000000e-01, 1.66351088e-02,
1.95919797e-01, 6.23358040e-02, 4.18740453e-03, 1.16061688e-03,
7.11297248e-03, 6.59679571e-03],
[5.94472678e-04, 1.33559792e-02, 1.66351088e-02, 2.00000000e-01,
1.59911246e-02, 6.43812362e-02, 5.90141166e-06, 6.77520700e-07,
1.52525053e-05, 1.33384349e-05],
[4.36813578e-02, 1.56539930e-01, 1.95919797e-01, 1.59911246e-02,
2.00000000e-01, 6.11287461e-02, 4.41158561e-03, 1.23488073e-03,
7.46433076e-03, 6.92846776e-03],
[7.54738292e-03, 5.58967934e-02, 6.23358040e-02, 6.43812362e-02,
6.11287461e-02, 2.00000000e-01, 2.30531400e-04, 4.11226399e-05,
4.89214341e-04, 4.42318453e-04],
[4.79816083e-02, 5.50622994e-03, 4.18740453e-03, 5.90141166e-06,
4.41158561e-03, 2.30531400e-04, 2.00000000e-01, 8.95609518e-02,
1.03946894e-01, 1.06810568e-01],
[2.44870899e-02, 1.61259456e-03, 1.16061688e-03, 6.77520700e-07,
1.23488073e-03, 4.11226399e-05, 8.95609518e-02, 2.00000000e-01,
7.89686728e-02, 8.05577562e-02],
[6.11804858e-02, 9.16550083e-03, 7.11297248e-03, 1.52525053e-05,
7.46433076e-03, 4.89214341e-04, 1.03946894e-01, 7.89686728e-02,
2.00000000e-01, 1.89352355e-01],
[5.91636643e-02, 8.54623880e-03, 6.59679571e-03, 1.33384349e-05,
6.92846776e-03, 4.42318453e-04, 1.06810568e-01, 8.05577562e-02,
1.89352355e-01, 2.00000000e-01]])
Conversion to ONNX#
The function is not an operator, the function to use is specific to this usage.
from skl2onnx.operator_converters.gaussian_process import convert_kernel
from skl2onnx.common.data_types import FloatTensorType, DoubleTensorType
from skl2onnx.algebra.onnx_ops import OnnxIdentity
onnx_op = convert_kernel(ker, 'X', output_names=['final_after_op_Add'],
dtype=numpy.float32, op_version=12)
onnx_op = OnnxIdentity(onnx_op, output_names=['Y'], op_version=12)
model_onnx = model_onnx = onnx_op.to_onnx(
inputs=[('X', FloatTensorType([None, None]))],
target_opset=12)
with open("model_onnx.onnx", "wb") as f:
f.write(model_onnx.SerializeToString())
[('X', FloatTensorType([None, None]))] means the function applies on
every tensor whatever its dimension is.
%onnxview model_onnx
from mlprodict.onnxrt import OnnxInference
from mlprodict.tools.asv_options_helper import get_ir_version_from_onnx
# line needed when onnx is more recent than onnxruntime
model_onnx.ir_version = get_ir_version_from_onnx()
pyrun = OnnxInference(model_onnx, inplace=False)
rtrun = OnnxInference(model_onnx, runtime="onnxruntime1")
pyres = pyrun.run({'X': Xtest.astype(numpy.float32)})
pyres
{'Y': array([[2.00000003e-01, 4.88993339e-02, 4.25047986e-02, 5.94472338e-04,
4.36813496e-02, 7.54737947e-03, 4.79816124e-02, 2.44870633e-02,
6.11804537e-02, 5.91636561e-02],
[4.88993339e-02, 2.00000003e-01, 1.41439855e-01, 1.33559676e-02,
1.56540006e-01, 5.58967553e-02, 5.50623611e-03, 1.61259342e-03,
9.16550029e-03, 8.54624342e-03],
[4.25047986e-02, 1.41439855e-01, 2.00000003e-01, 1.66351143e-02,
1.95919767e-01, 6.23358004e-02, 4.18740092e-03, 1.16061396e-03,
7.11296080e-03, 6.59678876e-03],
[5.94472338e-04, 1.33559676e-02, 1.66351143e-02, 2.00000003e-01,
1.59911271e-02, 6.43812567e-02, 5.90140644e-06, 6.77518699e-07,
1.52524681e-05, 1.33384119e-05],
[4.36813496e-02, 1.56540006e-01, 1.95919767e-01, 1.59911271e-02,
2.00000003e-01, 6.11287355e-02, 4.41158377e-03, 1.23487751e-03,
7.46431900e-03, 6.92846393e-03],
[7.54737947e-03, 5.58967553e-02, 6.23358004e-02, 6.43812567e-02,
6.11287355e-02, 2.00000003e-01, 2.30531194e-04, 4.11224828e-05,
4.89213213e-04, 4.42317745e-04],
[4.79816124e-02, 5.50623611e-03, 4.18740092e-03, 5.90140644e-06,
4.41158377e-03, 2.30531194e-04, 2.00000003e-01, 8.95609260e-02,
1.03946947e-01, 1.06810644e-01],
[2.44870633e-02, 1.61259342e-03, 1.16061396e-03, 6.77518699e-07,
1.23487751e-03, 4.11224828e-05, 8.95609260e-02, 2.00000003e-01,
7.89686665e-02, 8.05577263e-02],
[6.11804537e-02, 9.16550029e-03, 7.11296080e-03, 1.52524681e-05,
7.46431900e-03, 4.89213213e-04, 1.03946947e-01, 7.89686665e-02,
2.00000003e-01, 1.89352334e-01],
[5.91636561e-02, 8.54624342e-03, 6.59678876e-03, 1.33384119e-05,
6.92846393e-03, 4.42317745e-04, 1.06810644e-01, 8.05577263e-02,
1.89352334e-01, 2.00000003e-01]], dtype=float32)}
rtres = rtrun.run({'X': Xtest.astype(numpy.float32)})
rtres
{'Y': array([[2.00000003e-01, 4.88993339e-02, 4.25047986e-02, 5.94472338e-04,
4.36813496e-02, 7.54737947e-03, 4.79816124e-02, 2.44870633e-02,
6.11804537e-02, 5.91636561e-02],
[4.88993339e-02, 2.00000003e-01, 1.41439855e-01, 1.33559657e-02,
1.56540006e-01, 5.58967553e-02, 5.50623611e-03, 1.61259342e-03,
9.16550029e-03, 8.54624342e-03],
[4.25047986e-02, 1.41439855e-01, 2.00000003e-01, 1.66351143e-02,
1.95919767e-01, 6.23358078e-02, 4.18740092e-03, 1.16061396e-03,
7.11296080e-03, 6.59678876e-03],
[5.94472338e-04, 1.33559657e-02, 1.66351143e-02, 2.00000003e-01,
1.59911271e-02, 6.43812567e-02, 5.90140644e-06, 6.77518699e-07,
1.52524681e-05, 1.33384119e-05],
[4.36813496e-02, 1.56540006e-01, 1.95919767e-01, 1.59911271e-02,
2.00000003e-01, 6.11287355e-02, 4.41158377e-03, 1.23487751e-03,
7.46431900e-03, 6.92846393e-03],
[7.54737947e-03, 5.58967553e-02, 6.23358078e-02, 6.43812567e-02,
6.11287355e-02, 2.00000003e-01, 2.30531194e-04, 4.11224828e-05,
4.89213213e-04, 4.42317745e-04],
[4.79816124e-02, 5.50623611e-03, 4.18740092e-03, 5.90140644e-06,
4.41158377e-03, 2.30531194e-04, 2.00000003e-01, 8.95609260e-02,
1.03946947e-01, 1.06810644e-01],
[2.44870633e-02, 1.61259342e-03, 1.16061396e-03, 6.77518699e-07,
1.23487751e-03, 4.11224828e-05, 8.95609260e-02, 2.00000003e-01,
7.89686665e-02, 8.05577263e-02],
[6.11804537e-02, 9.16550029e-03, 7.11296080e-03, 1.52524681e-05,
7.46431900e-03, 4.89213213e-04, 1.03946947e-01, 7.89686665e-02,
2.00000003e-01, 1.89352334e-01],
[5.91636561e-02, 8.54624342e-03, 6.59678876e-03, 1.33384119e-05,
6.92846393e-03, 4.42317745e-04, 1.06810644e-01, 8.05577263e-02,
1.89352334e-01, 2.00000003e-01]], dtype=float32)}
from mlprodict.onnxrt.validate.validate_difference import measure_relative_difference
measure_relative_difference(pyres['Y'], rtres['Y'])
9.059079e-08
The last runtime uses the same runtime but with double instead of floats.
onnx_op_64 = convert_kernel(ker, 'X', output_names=['final_after_op_Add'],
dtype=numpy.float64, op_version=12)
onnx_op_64 = OnnxIdentity(onnx_op_64, output_names=['Y'], op_version=12)
model_onnx_64 = onnx_op_64.to_onnx(
inputs=[('X', DoubleTensorType([None, None]))],
target_opset=12)
pyrun64 = OnnxInference(model_onnx_64, runtime="python", inplace=False)
pyres64 = pyrun64.run({'X': Xtest.astype(numpy.float64)})
measure_relative_difference(pyres['Y'], pyres64['Y'])
7.106326595962827e-07
Side by side#
We run every node independently and we compare the output at each step.
%matplotlib inline
from mlprodict.onnxrt.validate.side_by_side import side_by_side_by_values
from pandas import DataFrame
def run_sbs(r1, r2, r3, x):
sbs = side_by_side_by_values([r1, r2, r3],
inputs=[
{'X': x.astype(numpy.float32)},
{'X': x.astype(numpy.float32)},
{'X': x.astype(numpy.float64)},
])
df = DataFrame(sbs)
dfd = df.drop(['value[0]', 'value[1]', 'value[2]'], axis=1).copy()
dfd.loc[dfd.cmp == 'ERROR->=inf', 'v[1]'] = 10
return dfd, sbs
dfd, _ = run_sbs(pyrun, rtrun, pyrun64, Xtest)
dfd
| metric | step | v[0] | v[1] | v[2] | cmp | name | shape[0] | shape[1] | shape[2] | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | nb_results | -1 | 49 | 4.900000e+01 | 4.900000e+01 | OK | NaN | NaN | NaN | NaN |
| 1 | abs-diff | 0 | 0 | 0.000000e+00 | 3.250289e-08 | OK | X | (10, 6) | (10, 6) | (10, 6) |
| 2 | abs-diff | 1 | 0 | 9.059079e-08 | 7.106327e-07 | OK | Y | (10, 10) | (10, 10) | (10, 10) |
| 3 | abs-diff | 2 | 0 | 0.000000e+00 | 1.490116e-08 | OK | Ad_Addcst | (1,) | (1,) | (1,) |
| 4 | abs-diff | 3 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Sh_shape0 | (2,) | (2,) | (2,) |
| 5 | abs-diff | 4 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Co_output0 | (10, 6) | (10, 6) | (10, 6) |
| 6 | abs-diff | 5 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Re_reduced0 | (10, 1) | (10, 1) | (10, 1) |
| 7 | abs-diff | 6 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Tr_transposed0 | (1, 10) | (1, 10) | (1, 10) |
| 8 | abs-diff | 7 | 0 | 1.000000e+00 | 7.106327e-07 | ERROR->=1.0 | Ma_Y0 | (10, 10) | (10, 10) | (10, 10) |
| 9 | abs-diff | 8 | 0 | 4.863269e+00 | 7.106327e-07 | ERROR->=4.9 | Ad_C0 | (10, 10) | (10, 10) | (10, 10) |
| 10 | abs-diff | 9 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Sh_shape02 | (2,) | (2,) | (2,) |
| 11 | abs-diff | 10 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Co_output02 | (10, 6) | (10, 6) | (10, 6) |
| 12 | abs-diff | 11 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Re_reduced01 | (6,) | (6,) | (6,) |
| 13 | abs-diff | 12 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Sh_shape01 | (1,) | (1,) | (1,) |
| 14 | abs-diff | 13 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Co_output01 | (6,) | (6,) | (6,) |
| 15 | abs-diff | 14 | 0 | 0.000000e+00 | 4.969472e-08 | OK | Di_C0 | (10, 6) | (10, 6) | (10, 6) |
| 16 | abs-diff | 15 | 0 | 0.000000e+00 | 4.969472e-08 | OK | scan0 | (10, 6) | (10, 6) | (10, 6) |
| 17 | abs-diff | 16 | 0 | 9.471974e+01 | 7.215496e-07 | ERROR->=94.7 | scan1 | (10, 10) | (10, 10) | (10, 10) |
| 18 | abs-diff | 17 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Sh_shape04 | (2,) | (2,) | (2,) |
| 19 | abs-diff | 18 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Co_output04 | (10, 6) | (10, 6) | (10, 6) |
| 20 | abs-diff | 19 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Re_reduced02 | (10, 1) | (10, 1) | (10, 1) |
| 21 | abs-diff | 20 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Sh_shape03 | (2,) | (2,) | (2,) |
| 22 | abs-diff | 21 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Co_output03 | (10, 1) | (10, 1) | (10, 1) |
| 23 | abs-diff | 22 | 0 | 4.736030e+01 | 7.215496e-07 | ERROR->=47.4 | Mu_C01 | (10, 10) | (10, 10) | (10, 10) |
| 24 | abs-diff | 23 | 0 | 7.247263e-08 | 7.215496e-07 | OK | Ex_output0 | (10, 10) | (10, 10) | (10, 10) |
| 25 | abs-diff | 24 | 0 | 5.000000e-01 | 7.106327e-07 | ERROR->=0.5 | Mu_C0 | (10, 10) | (10, 10) | (10, 10) |
| 26 | abs-diff | 25 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Sh_shape05 | (2,) | (2,) | (2,) |
| 27 | abs-diff | 26 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Co_output05 | (10, 6) | (10, 6) | (10, 6) |
| 28 | abs-diff | 27 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Re_reduced03 | (10, 1) | (10, 1) | (10, 1) |
| 29 | abs-diff | 28 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Tr_transposed01 | (1, 10) | (1, 10) | (1, 10) |
| 30 | abs-diff | 29 | 0 | 1.000000e+00 | 6.830406e-07 | ERROR->=1.0 | Ma_Y01 | (10, 10) | (10, 10) | (10, 10) |
| 31 | abs-diff | 30 | 0 | 0.000000e+00 | 1.490116e-08 | OK | Ad_Addcst1 | (1,) | (1,) | (1,) |
| 32 | abs-diff | 31 | 0 | 1.000000e+00 | 6.830406e-07 | ERROR->=1.0 | Ad_C01 | (10, 10) | (10, 10) | (10, 10) |
| 33 | abs-diff | 32 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Sh_shape07 | (2,) | (2,) | (2,) |
| 34 | abs-diff | 33 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Co_output07 | (10, 6) | (10, 6) | (10, 6) |
| 35 | abs-diff | 34 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Re_reduced04 | (6,) | (6,) | (6,) |
| 36 | abs-diff | 35 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Sh_shape06 | (1,) | (1,) | (1,) |
| 37 | abs-diff | 36 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Co_output06 | (6,) | (6,) | (6,) |
| 38 | abs-diff | 37 | 0 | 0.000000e+00 | 3.250289e-08 | OK | Di_C01 | (10, 6) | (10, 6) | (10, 6) |
| 39 | abs-diff | 38 | 0 | 0.000000e+00 | 3.250289e-08 | OK | scan01 | (10, 6) | (10, 6) | (10, 6) |
| 40 | abs-diff | 39 | 0 | 1.947547e+03 | 6.849032e-07 | ERROR->=1947.5 | scan11 | (10, 10) | (10, 10) | (10, 10) |
| 41 | abs-diff | 40 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Sh_shape09 | (2,) | (2,) | (2,) |
| 42 | abs-diff | 41 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Co_output09 | (10, 6) | (10, 6) | (10, 6) |
| 43 | abs-diff | 42 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Re_reduced05 | (10, 1) | (10, 1) | (10, 1) |
| 44 | abs-diff | 43 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Sh_shape08 | (2,) | (2,) | (2,) |
| 45 | abs-diff | 44 | 0 | 0.000000e+00 | 0.000000e+00 | OK | Co_output08 | (10, 1) | (10, 1) | (10, 1) |
| 46 | abs-diff | 45 | 0 | 9.737733e+02 | 6.849032e-07 | ERROR->=973.8 | Mu_C03 | (10, 10) | (10, 10) | (10, 10) |
| 47 | abs-diff | 46 | 0 | 0.000000e+00 | 6.849032e-07 | OK | Ex_output01 | (10, 10) | (10, 10) | (10, 10) |
| 48 | abs-diff | 47 | 0 | 0.000000e+00 | 6.830406e-07 | OK | Mu_C02 | (10, 10) | (10, 10) | (10, 10) |
| 49 | abs-diff | 48 | 0 | 9.059079e-08 | 7.106327e-07 | OK | final_after_op_Add | (10, 10) | (10, 10) | (10, 10) |
ax = dfd[['name', 'v[2]']].iloc[1:].set_index('name').plot(kind='bar', figsize=(14,4), logy=True)
ax.set_title("relative difference for each output between python and onnxruntime");
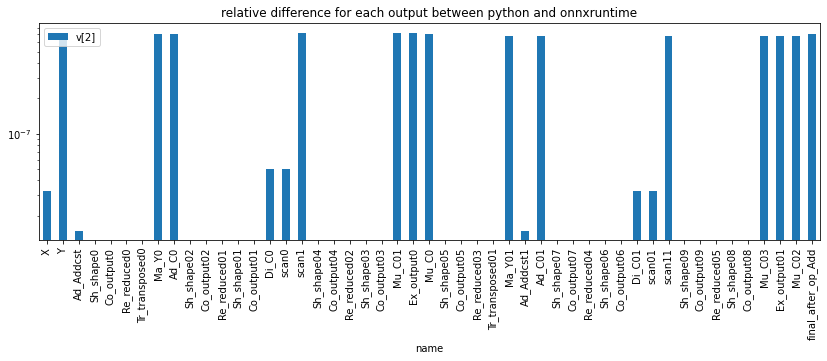
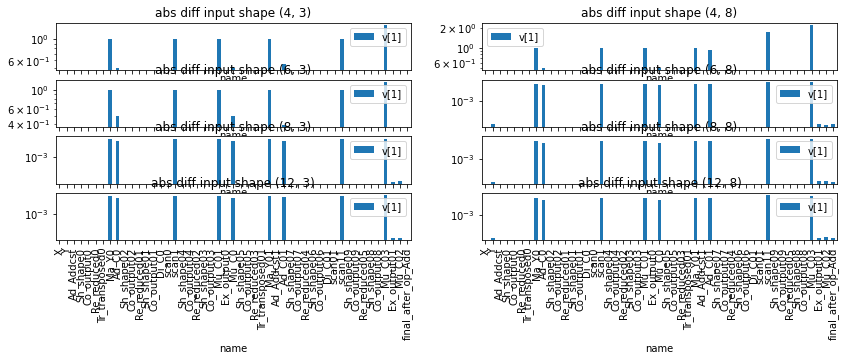
Let’s try for other inputs.
import warnings
from matplotlib.cbook.deprecation import MatplotlibDeprecationWarning
import matplotlib.pyplot as plt
with warnings.catch_warnings():
warnings.simplefilter("ignore", MatplotlibDeprecationWarning)
values = [4, 6, 8, 12]
fig, ax = plt.subplots(len(values), 2, figsize=(14, len(values) * 4))
for i, d in enumerate(values):
for j, dim in enumerate([3, 8]):
mat = numpy.random.rand(d, dim)
dfd, _ = run_sbs(pyrun, rtrun, pyrun64, mat)
dfd[['name', 'v[1]']].iloc[1:].set_index('name').plot(
kind='bar', figsize=(14,4), logy=True, ax=ax[i, j])
ax[i, j].set_title("abs diff input shape {}".format(mat.shape))
if i < len(values) - 1:
for xlabel_i in ax[i, j].get_xticklabels():
xlabel_i.set_visible(False)
Further analysis#
If there is one issue, we can create a simple graph to test. We consider
Y = A + B where A and B have the following name in the ONNX
graph:
node = pyrun.sequence_[-2].onnx_node
final_inputs = list(node.input)
final_inputs
['Mu_C0', 'Mu_C02']
_, sbs = run_sbs(pyrun, rtrun, pyrun64, Xtest)
names = final_inputs + ['Y']
values = {}
for row in sbs:
if row.get('name', '#') not in names:
continue
name = row['name']
values[name] = [row["value[%d]" % i] for i in range(3)]
list(values.keys())
['Y', 'Mu_C0', 'Mu_C02']
Let’s check.
for name in names:
if name not in values:
raise Exception("Unable to find '{}' in\n{}".format(
name, [_.get('name', "?") for _ in sbs]))
a, b, c = names
for i in [0, 1, 2]:
A = values[a][i]
B = values[b][i]
Y = values[c][i]
diff = Y - (A + B)
dabs = numpy.max(numpy.abs(diff))
print(i, diff.dtype, dabs)
0 float32 0.10000001
1 float32 0.0
2 float64 0.10000000000000003
If the second runtime has issue, we can create a single node to check something.
from skl2onnx.algebra.onnx_ops import OnnxAdd
onnx_add = OnnxAdd('X1', 'X2', output_names=['Y'], op_version=12)
add_onnx = onnx_add.to_onnx({'X1': A, 'X2': B}, target_opset=12)
add_onnx.ir_version = get_ir_version_from_onnx()
pyrun_add = OnnxInference(add_onnx, inplace=False)
rtrun_add = OnnxInference(add_onnx, runtime="onnxruntime1")
res1 = pyrun_add.run({'X1': A, 'X2': B})
res2 = rtrun_add.run({'X1': A, 'X2': B})
measure_relative_difference(res1['Y'], res2['Y'])
0.0
No mistake here.
onnxruntime#
from onnxruntime import InferenceSession, RunOptions, SessionOptions
opt = SessionOptions()
opt.enable_mem_pattern = True
opt.enable_cpu_mem_arena = True
sess = InferenceSession(model_onnx.SerializeToString(), opt)
sess
<onnxruntime.capi.session.InferenceSession at 0x22d1e846278>
res = sess.run(None, {'X': Xtest.astype(numpy.float32)})[0]
measure_relative_difference(pyres['Y'], res)
9.059079e-08
res = sess.run(None, {'X': Xtest.astype(numpy.float32)})[0]
measure_relative_difference(pyres['Y'], res)
9.059079e-08
Side by side for MLPRegressor#
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
from sklearn.neural_network import MLPRegressor
iris = load_iris()
X, y = iris.data, iris.target
X_train, X_test, y_train, y_test = train_test_split(X, y)
clr = MLPRegressor()
clr.fit(X_train, y_train)
MLPRegressor()
from mlprodict.onnx_conv import to_onnx
onx = to_onnx(clr, X_train.astype(numpy.float32), target_opset=12)
onx.ir_version = get_ir_version_from_onnx()
pyrun = OnnxInference(onx, runtime="python", inplace=False)
rtrun = OnnxInference(onx, runtime="onnxruntime1")
rt_partial_run = OnnxInference(onx, runtime="onnxruntime2")
dfd, _ = run_sbs(rtrun, rt_partial_run, pyrun, X_test)
dfd
| metric | step | v[0] | v[1] | v[2] | cmp | name | shape[0] | shape[1] | shape[2] | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | nb_results | -1 | 13 | 13.0 | 1.300000e+01 | OK | NaN | NaN | NaN | NaN |
| 1 | abs-diff | 0 | 0 | 0.0 | 3.973643e-08 | OK | X | (38, 4) | (38, 4) | (38, 4) |
| 2 | abs-diff | 1 | 0 | 0.0 | 0.000000e+00 | OK | coefficient | (4, 100) | (4, 100) | (4, 100) |
| 3 | abs-diff | 2 | 0 | 0.0 | 0.000000e+00 | OK | intercepts | (1, 100) | (1, 100) | (1, 100) |
| 4 | abs-diff | 3 | 0 | 0.0 | 0.000000e+00 | OK | coefficient1 | (100, 1) | (100, 1) | (100, 1) |
| 5 | abs-diff | 4 | 0 | 0.0 | 0.000000e+00 | OK | intercepts1 | (1, 1) | (1, 1) | (1, 1) |
| 6 | abs-diff | 5 | 0 | 0.0 | 0.000000e+00 | OK | shape_tensor | (2,) | (2,) | (2,) |
| 7 | abs-diff | 6 | 0 | 0.0 | 0.000000e+00 | OK | cast_input | (38, 4) | (38, 4) | (38, 4) |
| 8 | abs-diff | 7 | 0 | 0.0 | 1.000000e+00 | ERROR->=1.0 | mul_result | (38, 100) | (38, 100) | (38, 100) |
| 9 | abs-diff | 8 | 0 | 0.0 | 1.000000e+00 | ERROR->=1.0 | add_result | (38, 100) | (38, 100) | (38, 100) |
| 10 | abs-diff | 9 | 0 | 0.0 | 0.000000e+00 | OK | next_activations | (38, 100) | (38, 100) | (38, 100) |
| 11 | abs-diff | 10 | 0 | 0.0 | 2.311237e-02 | e<0.1 | mul_result1 | (38, 1) | (38, 1) | (38, 1) |
| 12 | abs-diff | 11 | 0 | 0.0 | 0.000000e+00 | OK | add_result1 | (38, 1) | (38, 1) | (38, 1) |
| 13 | abs-diff | 12 | 0 | 0.0 | 0.000000e+00 | OK | variable | (38, 1) | (38, 1) | (38, 1) |
%onnxview onx